The infrastructure of the Aquaculture Microbiology Lab is designed to support mechanistic, comparative, and experimental research on fish disease across aquatic systems.
Rather than being organized around isolated techniques, our facilities integrate microbiology, experimental infection models, large-scale aquaculture access, and bioprocessing capacities.
This integrated setup allows us to study microbial adaptation, host–pathogen interactions, and disease dynamics under controlled, semi-controlled, and applied conditions.
Biological collections and microbial resources
The lab maintains one of the most extensive collections of bacterial pathogens associated with marine aquaculture in the Eastern Mediterranean.
This collection comprises hundreds of bacterial isolates obtained from farmed and wild fish, primarily from marine environments.
Isolates are extensively characterized and constitute a core resource for epidemiological and comparative studies, analysis of pathogen diversity and population structure, experimental infection and virulence studies, and development and evaluation of prevention-oriented strategies.
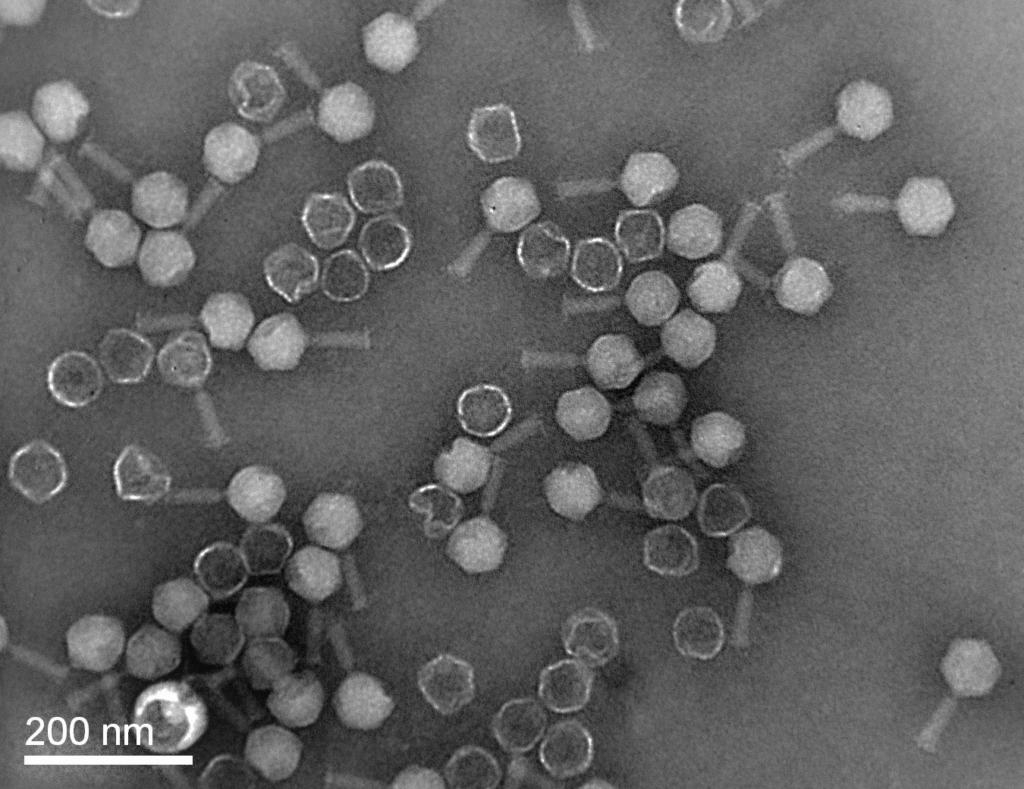
In parallel, the lab maintains a large and continuously expanding collection of bacteriophages, with particular emphasis on phages targeting aquaculture-relevant bacterial pathogens. Together, these collections provide a unique platform for integrated studies of pathogen adaptation and biological control.
Experimental infection models
To study disease processes under controlled conditions, the lab has developed innovative experimental infection platforms tailored to aquatic research.
A key model is based on fish larvae at the pre-feeding yolk-sac stage, which are exempt from animal experimentation regulations.
Larvae are individually housed in multiwell formats, enabling high-throughput assessment of pathogen virulence, infection dynamics, and strain competition.
This system allows quantitative analysis of disease outcome, comparison of pathogen variants and resistance phenotypes, and investigation of host–pathogen interactions in vivo.
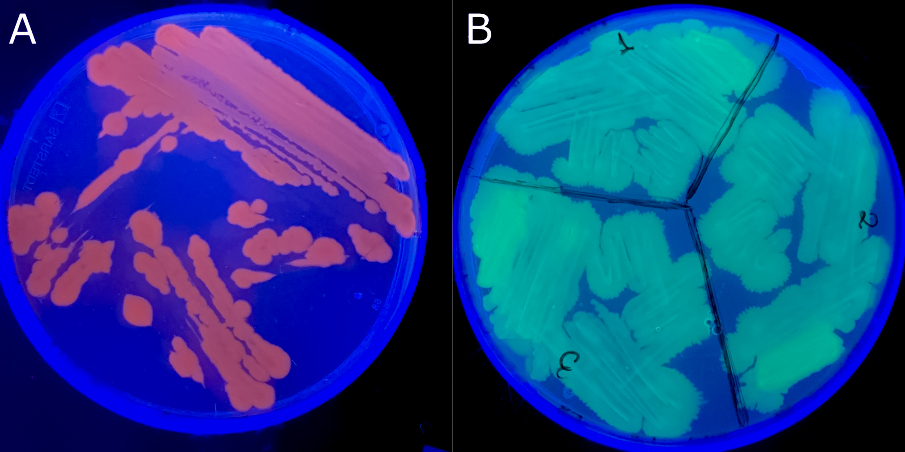
Fluorescently labelled pathogens and fluorescence microscopy
The lab has developed bacterial pathogens expressing fluorescent reporters (GFP and RFP), enabling direct visualization of infection processes in living hosts.
Combined with fluorescence microscopy, this approach supports tracking of pathogen colonization, tissue tropism, and competitive interactions in vivo.
These tools provide mechanistic insight into virulence expression and pathogen behavior that cannot be obtained through endpoint measurements alone.
Direct access to large-scale aquaculture facilities
The Aquaculture Microbiology Lab has direct access to the large-scale aquaculture facilities of the Institute of Marine Biology, Biotechnology and Aquaculture (IMBBC).
This proximity enables validation of laboratory findings under realistic aquaculture conditions, disease challenge and monitoring at production-relevant scales, and integration of experimental research with applied fish health management.
Local research environment and access to complementary facilities
The lab operates within a tightly connected local research ecosystem in Heraklion, providing direct access to complementary facilities at the University of Crete.
Through established collaborations, the lab has direct access to complementary facilities at the University of Crete, including the Fish Physiology Laboratory, the Electron Microscopy Laboratory, and the Sarris Laboratory, which supports bacterial engineering and genetic manipulation for mechanistic microbiology and experimental design.
The close physical proximity of these facilities allows seamless integration into ongoing research workflows, functioning as an extension of the lab’s core infrastructure.
Microbiology and growth analysis infrastructure
The Aquaculture Microbiology Lab is fully equipped for classical and advanced microbiological work, including biosafety level II laminar flow cabinets, controlled-temperature incubators, autoclaves, centrifuges, and cold storage facilities.
Bacterial growth and physiology are monitored using incubating multiwell photometric systems, enabling real-time analysis of growth kinetics and responses to biological and chemical stressors.
Bioprocessing and scale-up facilities
To bridge laboratory-scale experiments with applied research, the lab operates bioprocessing infrastructure that supports controlled microbial cultivation and scale-up.
Facilities include parallel small-scale bioreactors for replicated experimental designs and medium-scale bioreactors for bacterial biomass and bacteriophage production.
Ongoing upgrades further expand capacity for concentration, purification, and downstream processing, supporting translational research and prevention-oriented applications.

Infrastructure in perspective
The facilities of the Aquaculture Microbiology Lab are not an end in themselves, but a means to support coherent, hypothesis-driven research on fish disease.
By combining in-house infrastructure with direct access to aquaculture facilities and closely integrated local laboratories, the lab maintains the flexibility to move seamlessly between fundamental research and applied validation.
This integrated infrastructure supports long-term efforts to reduce antibiotic dependence in aquaculture by enabling prevention-oriented strategies based on targeted and precise interventions, as well as tailored disease control approaches.
